Brian Wade D. Jamandre1, Apolinario V. Yambot1, Kang Ning Shen2 and Wann Nian Tzeng2
1College of Fisheries – Freshwater Aquaculture Center, Central Luzon State University, Science City of Muñoz, Nueva Ecija 3120, Philippines
2Department of Zoology and Institute of Fisheries Science,College of Life Science, National Taiwan University, Taipei, Taiwan ROC

Freshwater eels Anguilla spp are catadromous fishes migrating between freshwater and marine environment. The eels have a multi-stage life cycle and spawn in the open ocean. After spawning, their leaf-like larvae, leptocephali, drift with the oceanic currents towards their continental growth habitat. The leptocephali metamorphose to glass eels in the continental shelf and develop to elvers when they reach the estuary. The elvers eventually settle in the brackish coastal water, estuaries and rivers to grow into yellow eels. During maturation, they transform to silver eels and go back to the oceanic spawning ground to reproduce and die.
Most of the 18 species of anguillid eels in the world are tropical species and reside in the Indo-Pacific region. The Philippine archipelago lies within the region near the major spawning grounds of the tropical eels in the western waters of the Marianas Islands. With the influence of the North Equatorial Current and archipelagic nature, many freshwater eels thrive in the Philippine waters.
Differentiating eel species based on morphology encounters difficulty because of similarity in external features. Hence, species identification and evolutionary relationship based on molecular genetics are relatively reliable.
In the Philippines, freshwater eels have different populations scattered in various regions in country in which they are considered important food. However, there has been no document on the evolutionary history and phylogenetic studies of the eels in the country either by traditional or molecular methods. This study therefore aimed to identify the eel species and determine their phylogenetic relationship by analyzing the 16S rRNA and cytochrome b genes of the mitochondrial DNA.
Table 1. Primers used in the amplification of the 16S rRNA and cytochrome b of Philippine freshwater eels.
| A. Cytochrome b gene | |
| H15341 | 5’ – TGC TAA CGA CCT AGT GG – 3’ |
| L151341 | 5’ – CTA GTC AAC CTA CTA ATG GG – 3’ |
| B. 16S rRNA gene | |
| L1374 | 5’ – GAA GAA ATG GGC TAC ATT TTC TA – 3’ |
| H2009 | 5’ – CCT AAG CAA CCA GCT ATA AC – 3’ |
| L1854 | 5’ – AAA CCT CGT ACC TTT TGC AT – 3’ |
| H2582 | 5’ – ATT GCG CTA CCT TTG CAC GGT – 3’ |
Eels were collected from the Cagayan River system and Chico River in the northern Luzon, Philippines. DNA extraction from muscle tissues of the collected eel samples, PCR amplification and sequencing were carried out using the primers listed in Table 1. The 16S rRNA and cytochrome b genes of the mtDNA of the eels were sequenced and the nucleotides consisted of 813 and 920 base pairs, respectively. Nucleotide sequences were aligned and phylogenetic trees were constructed using DAMBE and MEGA2.
Pairwise comparisons, sequence alignment, and DNA analysis of the 16S rRNA and cytochrome b genes partial sequences of Philippine eels showed low values of nucleotide differences within species. This indicates that there are only little pressures (e.g. fishing pressures, exploitation, natural barriers) influencing the population of the Philippine eels making the sequences of these genes conserved among the three species. In addition, DNA sequences revealed that three eel species comprised the collected samples namely Anguilla marmorata, A. bicolor bicolor, and A. bicolor pacifica (Figs. 1, 2 & 3). The 16S rRNA phylogenetic tree showed that the samples from the Cagayan River system formed a clade with the A. bicolor pacifica whereas the samples from the Chico river formed a clade with A. marmorata, with robust bootstrap support. The same results were also obtained with the cytochrome b gene, only that some samples from the Cagayan River formed a clade with A. bicolor bicolor. Phylogenetic analysis further indicates that the Philippine freshwater eels belong to the Indo-Pacific lineage. A. marmorata was the most basal (ancestral) species of the lineage among the three species suggesting an Indo-Malayan origin and A. bicolor was the recently evolved species. This study is the first record of molecular characterization of freshwater eels in the Philippines.
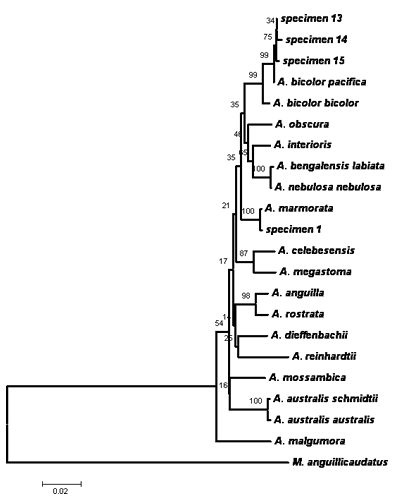 |
| Figure 1. Phylogenetic tree inferred from 16S rRNA gene using neighbor-joining method. Specimen 13, 14 and 15 (identified as Anguilla bicolor pacifica) were collected from the Cagayan River system and specimen 1 (A. marmorata) was collected from Chico River. |
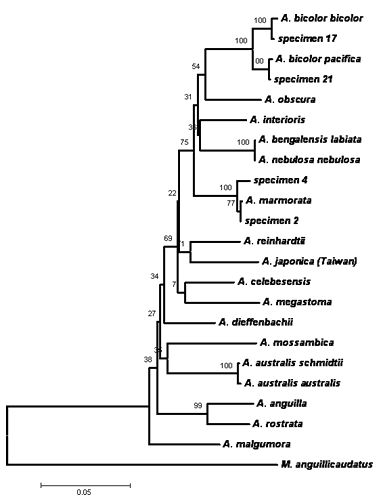 |
| Figure 2. Phylogenetic tree inferred from the cytochrome b coding region of the mtDNA using neighbor-joining method. Specimen 17 (identified as Anguilla bicolor bicolor) and specimen 21 (A. bicolor pacifica) were collected from the Cagayan River System, and specimen 2 and 4 (A. marmorata) were collected from Chico River. |
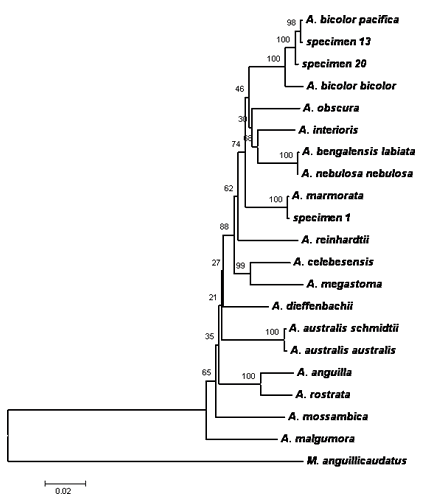 |
| Figure 3. Phylogenetic tree inferred from the combined cytochrome b and 16S rRNA gene sequence data. Specimen 13 and 20 (identified as Anguilla bicolor pacifica) were collected from the Cagayan River system and specimen 1 (A. marmorata) was collected from Chico River. |
Reference:
Jamandre, BWD, AV Yambot, KN Shen, WN Tzeng. 2007. Molecular phylogeny of Philippine freshwater eels Anguilla spp (Actinopterygi: Anguilliformes: Anguilidae) inferred from mitochondrial DNA. Raffles Bulletin of Zoology 14:43-51.
__________
Dr. Apolinario V. Yambot is the current dean of the College of Fisheries, Central Luzon State University in Munoz, Nueva Ecija. He obtained his Ph.D. (specialization in fish immunology) from the National Taiwan University, Taipei, Taiwan, ROC. His research interests include vaccine, probiotics, cortisol response, DNA characterization, otolith microchemistry.